Bayesian Estimation by using rjags Package
JAGS is the abbreviation for Just Another Gibbs Sampler which is used for easy Bayesian modeling. rjags is a kind of wrapper of jags. This post will use rjags R package to estimate a multiple linear regression model by Bayesian MCMC. Prior to installing rjags, jags should be installed at first.
Likelihood
The likelihood of a multiple linear regression model (k=2) is constructed from the following model specification. \((t=1,2,…,n)\)
\[\begin{align} Y_t &\sim \text{Normal}(\mu, \tau) \\ \mu &= \beta_0+\beta_1 X_{1t}+\beta_2 X_{2t} \\ \sigma &= 1/\sqrt{\tau} \end{align}\]
Here, \(\tau\) in \(\text{Normal}(\mu, \tau)\) is not a variance but a precision which is the reciprocal of a variance (\(\tau = \frac{1}{\sigma^2}\)).
Prior
Priors for regression coefficients and the reciprocal of an error variance should be given. Although there is no 'true' prior distribution, there is a 'satisficing' prior distribution. It can be done by a lot of experience or from the previous studies.
In our case, \(\beta\) are assumed to have normal priors but \(\tau\) is non-negative and is assumed to have a gamma prior. We use \(\hat{\beta_0}, \hat{\beta_1}, \hat{\beta_2}\) which are OLS estimates as each mean of priors for regression coefficients. Precision is set to 0.01 to represent our confidence is moderate and is not strong.
\[\begin{align} \beta_0 &\sim \text{Normal}(\hat{\beta_0}, 0.01) \\ \beta_1 &\sim \text{Normal}(\hat{\beta_1}, 0.01) \\ \beta_2 &\sim \text{Normal}(\hat{\beta_2}, 0.01) \\ \tau &\sim \text{Gamma}(0.01, 0.01) \end{align}\]
To use rjags, we have only to define model and each prior for coefficients without any lengthy derivations.
Model String in rjags
In rjags, a main part is a model string which consists of a likelihood and priors (with or without transformations). It is the same as the above equations essentially. When our model (likelihood) or priors are changed, we need to modify this model string properly.
R code
As an example, we take an AR(2) model as a test model. Data is the U.S. quarterly GDP growth rate (yoy) ranging from 1947Q1 to 2019Q1. In the next R code, jags.script is the model string.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 | #========================================================# # Quantitative Financial Econometrics & Derivatives # ML/DL using R, Python, Tensorflow by Sang-Heon Lee # # https://shleeai.blogyield.com #--------------------------------------------------------# # rjags example for Bayesian multiple regression #========================================================# graphics.off(); rm(list = ls()) library(rjags) library(coda) #----------------------------------------------------- # read data (U.S. quarterly GDP growth rate year-on-year) #----------------------------------------------------- # quarterly data for U.S. GDP growth rate (yoy) gdp_yoy <-c( 0.0257139670, 0.0447562683, 0.0525359910, 0.0381317790, 0.0093259088, -0.0104627119, -0.0059041116, -0.0154490719, 0.0369748307, 0.0704341513, 0.0980788417, 0.1254736302, 0.1004152961, 0.0875517208, 0.0700361239, 0.0532694875, 0.0504080513, 0.0353806505, 0.0221731143, 0.0522873341, 0.0600764198, 0.0656249942, 0.0527893859, 0.0052196433, -0.0179898299, -0.0246006338, -0.0077274628, 0.0269059373, 0.0598597082, 0.0749112146, 0.0770952717, 0.0637027107, 0.0316489075, 0.0237374609, 0.0094243285, 0.0197643323, 0.0300394541, 0.0196166557, 0.0302665177, 0.0035414395, -0.0291434042, -0.0203977878, -0.0072740159, 0.0262493878, 0.0715814738, 0.0873426752, 0.0651795264, 0.0449022299, 0.0480956652, 0.0203738001, 0.0245428618, 0.0087791174, -0.0067174527, 0.0155233387, 0.0296569900, 0.0619956746, 0.0729468622, 0.0651106689, 0.0583064298, 0.0421683677, 0.0353464754, 0.0375090926, 0.0470449534, 0.0502963040, 0.0603030182, 0.0599724792, 0.0537286165, 0.0502782228, 0.0533241329, 0.0550495076, 0.0615364189, 0.0812312602, 0.0813724980, 0.0722292596, 0.0586735114, 0.0440647570, 0.0288308733, 0.0260330499, 0.0270165087, 0.0263581712, 0.0377310270, 0.0536885532, 0.0519915518, 0.0484049473, 0.0437450686, 0.0302104821, 0.0290740384, 0.0202599426, 0.0032441293, 0.0016268587, 0.0042170031, -0.0016684843, 0.0266150980, 0.0305934387, 0.0296160568, 0.0427453445, 0.0341702075, 0.0512175143, 0.0524209432, 0.0666718926, 0.0728962552, 0.0612801001, 0.0466076021, 0.0394491207, 0.0063674420, -0.0020862905, -0.0063096723, -0.0196461231, -0.0232590023, -0.0185129237, 0.0079550988, 0.0252192903, 0.0597054841, 0.0598956257, 0.0483960934, 0.0422431855, 0.0317503644, 0.0436947211, 0.0561051726, 0.0489047254, 0.0403457494, 0.0590058015, 0.0511281571, 0.0644582144, 0.0630677430, 0.0262218823, 0.0236181481, 0.0127638331, 0.0141078278, -0.0077808984, -0.0163713694, -0.0003914969, 0.0158731684, 0.0292547991, 0.0423481003, 0.0129153625, -0.0221468644, -0.0101568967, -0.0258912833, -0.0145366869, 0.0142131192, 0.0321625248, 0.0557863013, 0.0760345142, 0.0823011467, 0.0769304925, 0.0667314613, 0.0542590241, 0.0445441428, 0.0361798193, 0.0417411642, 0.0409714759, 0.0406189998, 0.0363467080, 0.0307157148, 0.0286666689, 0.0267990197, 0.0330364855, 0.0321503663, 0.0438187584, 0.0415476345, 0.0438695504, 0.0410730401, 0.0372852235, 0.0422469583, 0.0367975587, 0.0383368666, 0.0270684337, 0.0278223606, 0.0238411639, 0.0171246753, 0.0060106594, -0.0095473822, -0.0054029478, -0.0010277132, 0.0115967019, 0.0281855548, 0.0312037714, 0.0359977334, 0.0428964179, 0.0326695847, 0.0276917220, 0.0226193094, 0.0257511647, 0.0337339603, 0.0413861313, 0.0424524751, 0.0403331370, 0.0342190216, 0.0237385166, 0.0263811775, 0.0217583082, 0.0256785859, 0.0392405346, 0.0396996610, 0.0432626787, 0.0422360126, 0.0421785102, 0.0456794827, 0.0439007010, 0.0474109873, 0.0401430518, 0.0401600330, 0.0476381748, 0.0471132023, 0.0455598811, 0.0461236331, 0.0469470314, 0.0411378962, 0.0516217468, 0.0399447798, 0.0293013196, 0.0228343952, 0.0105169266, 0.0050232471, 0.0015336818, 0.0130966573, 0.0133071719, 0.0219020499, 0.0207290678, 0.0175542787, 0.0200810319, 0.0324846821, 0.0423539093, 0.0421448528, 0.0411709185, 0.0337402858, 0.0322909355, 0.0379764716, 0.0349895931, 0.0344545645, 0.0307824359, 0.0329887855, 0.0307180626, 0.0233893470, 0.0255771428, 0.0147141403, 0.0180922467, 0.0219651225, 0.0195425692, 0.0114268306, 0.0108650373, 0.0000187657, -0.0279169036, -0.0334437760, -0.0400352950, -0.0309725345, 0.0018270714, 0.0169579334, 0.0275769043, 0.0312874776, 0.0253699929, 0.0191222715, 0.0170685309, 0.0094456342, 0.0159653298, 0.0261720704, 0.0233405632, 0.0249669608, 0.0145788777, 0.0155972048, 0.0125376209, 0.0189926726, 0.0258053859, 0.0141565704, 0.0263693756, 0.0307011415, 0.0283625033, 0.0406403469, 0.0339376821, 0.0254217778, 0.0214119574, 0.0176133228, 0.0139843073, 0.0157940174, 0.0204649433, 0.0204575407, 0.0216035638, 0.0234362726, 0.0266744034, 0.0303073105, 0.0327109848, 0.0306872928, 0.0244537329, 0.0224051369) ndata <- length(gdp_yoy) # Y, Y(-1) Y(-2) df.yx <- data.frame(GDP = gdp_yoy[3:ndata], GDP_lag1 = gdp_yoy[2:(ndata-1)], GDP_lag2 = gdp_yoy[1:(ndata-2)]) #----------------------------------------------------- # regression by using lm #----------------------------------------------------- lm_est <- lm(GDP ~ ., data = df.yx) #----------------------------------------------------- # Bayesian regression by using rjags #----------------------------------------------------- # Prepare data and additional information(n) lt.Data <- list(x = df.yx[,-1], y = df.yx[,1], n = nrow(df.yx)) # for use additional information(b_guess) inside model lt.Data$b_guess <- lm_est$coefficients # Model string jags.script <- "#-------------------------------------------------- model{ # likelihood for( i in 1:n) { y[i] ~ dnorm(mu[i], tau) mu[i] <- beta0 +beta1*x[i,1] + beta2*x[i,2] } # priors beta0 ~ dnorm(b_guess[1], 0.1) beta1 ~ dnorm(b_guess[2], 0.1) beta2 ~ dnorm(b_guess[3], 0.1) tau ~ dgamma(.01,.01) # transform sigma <- 1 / sqrt(tau) }#-------------------------------------------------- " # compile mod1 <- jags.model(textConnection(jags.script), data = lt.Data, n.chains = 4, n.adapt = 1000) # run update(mod1, 4000) # posterior sampling mod1.samples <- coda.samples( model = mod1, variable.names = c("beta0", "beta1", "beta2", "sigma"), n.iter = 2000) # plot trace and posterior density for each parameter x11(); plot(mod1.samples) # descriptive statistics of posterior densities sm <- summary(mod1.samples); sm # correlation cor(mod1.samples[[1]]) #----------------------------------------------------- # mean parameters #----------------------------------------------------- # means of posteriors of parameters beta0.post.mean <- sm$statistics["beta0", "Mean"] beta1.post.mean <- sm$statistics["beta1", "Mean"] beta2.post.mean <- sm$statistics["beta2", "Mean"] sigma.post.mean <- sm$statistics["sigma", "Mean"] beta_post <- rbind(beta0.post.mean, beta1.post.mean, beta2.post.mean, sigma.post.mean) # real data and fitted values ypred <- as.matrix(cbind(1,df.yx[,-1]))%*%beta_post[-4] x11(width = 16/2, height = 9/2) matplot(cbind(ypred,df.yx[,1]), type=c("l","p"), pch = 16, lty=c(1,1), lwd=3, col=c("blue", "green")) # Frequentist and Bayesian coefficients coefs <- rbind(c(lm_est$coefficients, sigma(lm_est)), t(beta_post)) colnames(coefs) <- c("const", "beta1", "beta2", "sigma") rownames(coefs) <- c("Frequentist", "Bayesian") coefs | cs |
Except for jags.script, the remaining part is not changed. In particular, it is worth noting that variables in jags.script should be in lt.Data like
lt.Data$b_guess <- lm_est$coefficients | cs |
Therefore when additional variables should be used in model string (jags.script), we should add that variables into that data list (lt.Data).
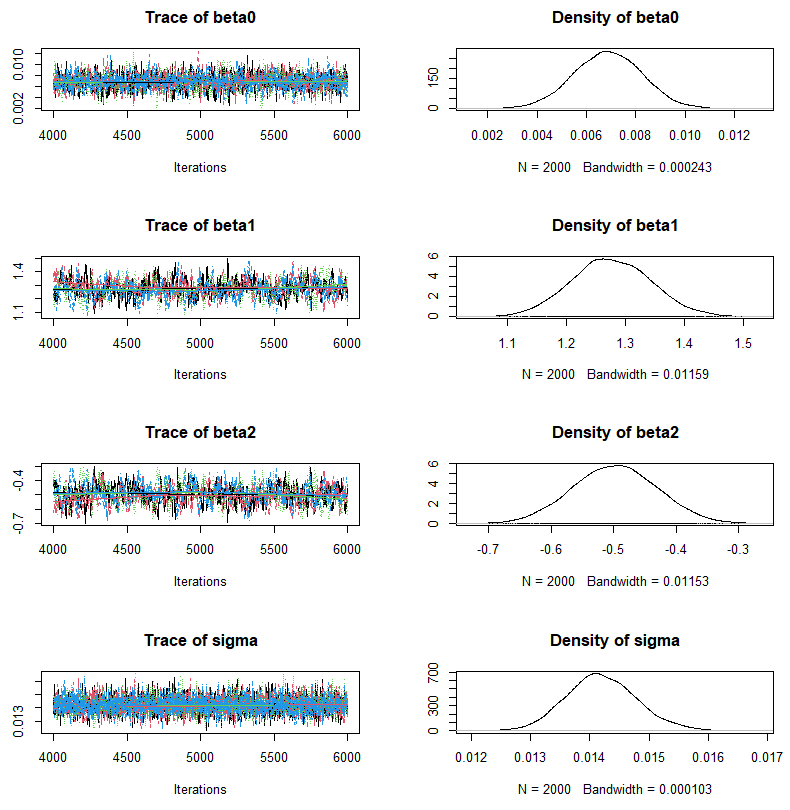
Estimation results contain of the following posterior distributions for each parameters.
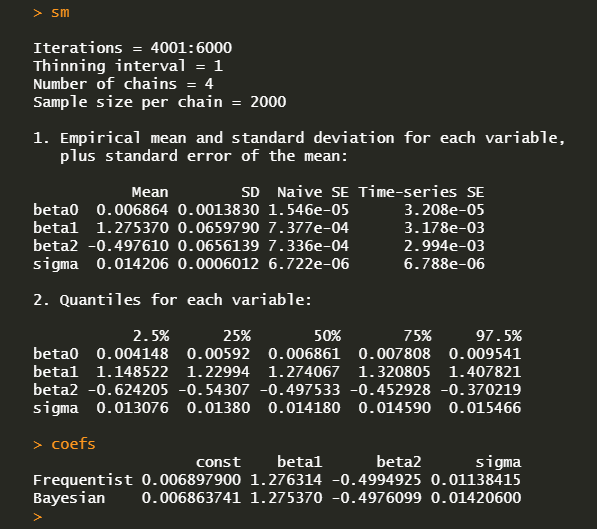
A summary statistics and average of posteriors can be obtained in the following way.
We can generate a time series of fitted response variable with average coefficients.
Concluding Remarks
This post shows how to use rjags R package to perform Bayesian MCMC estimation with multiple linear regression example. This is an easy tool for Bayesian analysis for us only if we understand the meaning of hyper-parameters of various prior distributions and use them without hesitation. \(\blacksquare\)

Won't logs be taken of negative numbers at times when dividing gdp rates? The data I am looking at may differ from the data in the example.
ReplyDeleteTaking into account your opinion, I pasted GDP growth rate data into R code directly for ease of use. Now there is no log in the data preparation stage.
DeleteGood luck.