Lasso, Group Lasso, and Exclusive Lasso
While LASSO (least absolute shrinkage and selection operator) has many variants and extensions, our focus is on two lasso models: Group Lasso and Exclusive Lasso. Before we dive into the specifics, let's go over the similarities and differences of these two lasso variants from the following figure.
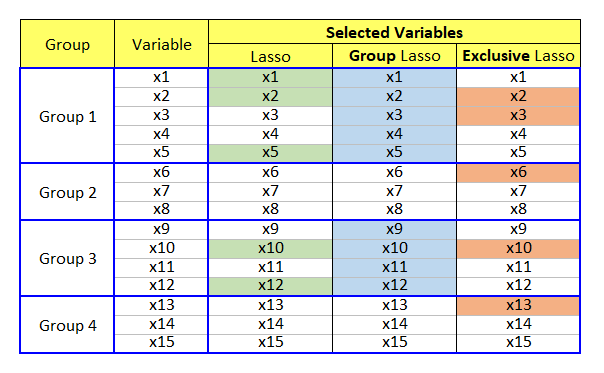
In the above figure, 15 variables are categorized into four groups. Lasso selects important features irrespective of the grouping. Of course, lasso did not select Group 2 and 4's variables but it is not intended but just an estimation result. While group lasso selects all or none in specific group, exclusive lasso selects at least one variable in each group.
From a perspective of competition, group lasso implements a completion across groups and on the contrary, exclusive lasso makes variables in the same group compete with each other within each group.
Since we can grasp the main characteristics of two lasso modes from the above figure, let's turn to the mathematical expressions.
Equations
There are some various expressions for these models and the next equations are for lasso, group lasso, and exclusive Lasso following Qiu et al. (2021).
\[\begin{align} \text{Lasso} &: \min_{\beta} \left\Vert y-X\beta \right\Vert_2^2 + \lambda \sum_{j=1}^{m} |\beta_j| \\ \text{Group Lasso} &: \min_{\beta} \left\Vert y-X\beta \right\Vert_2^2 + \lambda \sum_{g=1}^{G} \Vert \beta_g \Vert_2^1 \\ \text{Exclusive Lasso} &: \min_{\beta} \left\Vert y-X\beta \right\Vert_2^2 + \lambda \sum_{g=1}^{G} \Vert \beta_g \Vert_1^2 \end{align}\]
where the coefficient in \(\beta \) are divided into \(G\) groups and \(\beta_g\) denotes the coefficient vector of the \(g\)-th group.
In the group lasso, \(l_{2,1}\)-norm consists of the intra-group non-sparsity via \(l_2\)-norm and inter-group sparsity via \(l_1\)-norm. Therefore, variables of each group will be either selected or discarded entirely. Refer to Yuan and Lin (2006) for more information on the group lasso.
In exclusive lasso, \(l_{1,2}\)-norm consists of the intra-group sparsity via \(l_1\)-norm and inter-group non-sparsity via \(l_2\)-norm. Exclusive lasso selects at least one variable from each group. Refer to Zhou et al. (2010) for more information on the exclusive lasso.
R code
The following R code implements lasso, group lasso, and exclusive lasso for an artificial data set with a given group index. Required R packages are glmnet for lasso, gglasso for group lasso, and ExclusiveLasso for exclusive lasso.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 | #========================================================# # Quantitative Financial Econometrics & Derivatives # ML/DL using R, Python, Tensorflow by Sang-Heon Lee # # https://shleeai.blogspot.com #--------------------------------------------------------# # Group Lasso and Exclusive Lasso #========================================================# library(glmnet) library(gglasso) library(ExclusiveLasso) graphics.off() # clear all graphs rm(list = ls()) # remove all files from your workspace set.seed(1234) #-------------------------------------------- # X and y variable #-------------------------------------------- N = 500 # number of observations p = 20 # number of variables # random generated X X = matrix(rnorm(N*p), ncol=p) # standardization : mean = 0, std=1 X = scale(X) # artificial coefficients beta = c(0.15,-0.33,0.25,-0.25,0.05,0,0,0,0.5,0.2, -0.25, 0.12,-0.125,0,0,0,0,0,0,0) # Y variable, standardized Y y = X%*%beta + rnorm(N, sd=0.5) #y = scale(y) # group index for X variables v.group <- c(1,1,1,1,1,2,2,2,2,2, 3,3,3,3,3,4,4,4,4,4) #-------------------------------------------- # Model with a given lambda #-------------------------------------------- # lasso la <- glmnet(X, y, lambda = 0.1, family="gaussian", alpha=1, intercept = F) # group lasso gr <- gglasso(X, y, lambda = 0.2, group = v.group, loss="ls", intercept = F) # exclusive lasso ex <- exclusive_lasso(X, y,lambda = 0.2, groups = v.group, family="gaussian", intercept = F) # Results df.comp <- data.frame( group = v.group, beta = beta, Lasso = la$beta[,1], Group = gr$beta[,1], Exclusive = ex$coef[,1] ) df.comp #------------------------------------------------ # Run cross-validation & select lambda #------------------------------------------------ # lambda.min : minimal MSE # lambda.1se : the largest λ at which the MSE is # within one standard error of the minimal MSE. # lasso la_cv <- cv.glmnet(x=X, y=y, family='gaussian', alpha=1, intercept = F, nfolds=5) x11(); plot(la_cv) paste(la_cv$lambda.min, la_cv$lambda.1se) # group lasso gr_cv <- cv.gglasso(x=X, y=y, group=v.group, loss="ls", pred.loss="L2", intercept = F, nfolds=5) x11(); plot(gr_cv) paste(gr_cv$lambda.min, gr_cv$lambda.1se) # exclusive lasso ex_cv <- cv.exclusive_lasso( X, y, groups = v.group, intercept = F, nfolds=5) x11(); plot(ex_cv) paste(ex_cv$lambda.min, ex_cv$lambda.1se) #-------------------------------------------- # Model with selected lambda #-------------------------------------------- # lasso la <- glmnet(X, y, lambda = la_cv$lambda.1se, family="gaussian", alpha=1, intercept = F) # group lasso gr <- gglasso(X, y, lambda = gr_cv$lambda.1se+0.1, group = v.group, loss="ls", intercept = F) # exclusive lasso ex <- exclusive_lasso(X, y,lambda = ex_cv$lambda.1se, groups = v.group, family="gaussian", intercept = F) # Results df.comp.lambda.1se <- data.frame( group = v.group, beta = beta, Lasso = la$beta[,1], Group = gr$beta[,1], Exclusive = ex$coef[,1] ) df.comp.lambda.1se |
The first output from the above R code is the table of coefficients of all models with given each initial \(\lambda\) parameter. We can easily find the model-specific pattern of each model. I add horizontal dotted lines for separating each group just for exposition purpose.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 | > df.comp group beta Lasso Group Exclusive V1 1 0.150 0.01931728 0.016938769 0.013555753 V2 1 -0.330 -0.18832916 -0.047695924 -0.184065967 V3 1 0.250 0.17261562 0.042254702 0.169516525 V4 1 -0.250 -0.16322025 -0.043994211 -0.153730137 V5 1 0.050 0.00000000 0.009673207 0.000000000 -------------------------------------------------------- V6 2 0.000 0.00000000 0.001067915 0.000000000 V7 2 0.000 0.00000000 0.001355834 0.000000000 V8 2 0.000 0.00000000 0.014211932 0.000000000 V9 2 0.500 0.38757370 0.101900169 0.385382905 V10 2 0.200 0.11146785 0.044591933 0.110731304 -------------------------------------------------------- V11 3 -0.250 -0.15010738 0.000000000 -0.186626541 V12 3 0.120 0.00000000 0.000000000 0.003117881 V13 3 -0.125 -0.08305582 0.000000000 -0.120458426 V14 3 0.000 0.00000000 0.000000000 0.000000000 V15 3 0.000 0.00000000 0.000000000 0.000000000 -------------------------------------------------------- V16 4 0.000 0.00000000 0.000000000 0.000000000 V17 4 0.000 0.00000000 0.000000000 0.000000000 V18 4 0.000 0.00000000 0.000000000 0.010918904 V19 4 0.000 0.00000000 0.000000000 0.015330520 V20 4 0.000 0.00000000 0.000000000 0.013591628 |
The second output is the table of coefficients of all models with each selected lambda which is a result of cross validation. I add horizontal dotted lines for separating each group just for exposition purpose.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 | > df.comp.lambda.1se group beta Lasso Group Exclusive V1 1 0.150 0.07776181 4.779605e-02 0.08297141 V2 1 -0.330 -0.24670209 -1.257863e-01 -0.25235238 V3 1 0.250 0.22825029 1.130749e-01 0.23282822 V4 1 -0.250 -0.21384666 -1.168170e-01 -0.21582154 V5 1 0.050 0.03733144 2.717197e-02 0.04139150 ------------------------------------------------------- V6 2 0.000 0.00000000 2.184575e-03 0.00000000 V7 2 0.000 0.00000000 3.353260e-03 0.00000000 V8 2 0.000 0.01031027 2.950791e-02 0.02043597 V9 2 0.500 0.43538564 2.200230e-01 0.44419164 V10 2 0.200 0.16649806 9.620757e-02 0.17844727 ------------------------------------------------------- V11 3 -0.250 -0.20316169 -1.886308e-02 -0.22419384 V12 3 0.120 0.03113405 4.430739e-03 0.05392923 V13 3 -0.125 -0.13474237 -1.468172e-02 -0.15506193 V14 3 0.000 0.00000000 -3.646683e-05 0.00000000 V15 3 0.000 0.00000000 -1.311539e-03 0.00000000 ------------------------------------------------------- V16 4 0.000 0.00000000 0.000000e+00 0.00000000 V17 4 0.000 0.00000000 0.000000e+00 -0.01087451 V18 4 0.000 0.00000000 0.000000e+00 0.00000000 V19 4 0.000 0.00000000 0.000000e+00 0.01946948 V20 4 0.000 0.00000000 0.000000e+00 0.01318269 |
An Interesting Property of Exclusive Lasso
As stated earlier, the exclusive lasso selects at least one variable from each group. Let's check if this argument holds true with the next R code by setting \(\lambda\) to a higher value (100), which prevents from selecting variables.
1 2 3 4 5 6 7 8 9 10 11 12 13 | # lasso la <- glmnet(X, y, lambda = 100, family="gaussian", alpha=1, intercept = F) # group lasso gr <- gglasso(X, y, lambda = 100, group = v.group, loss="ls", intercept = F) # exclusive lasso ex <- exclusive_lasso(X, y,lambda = 100, groups = v.group, family="gaussian", intercept = F) |
The following result is sufficient for supporting the above explanation. While lasso and group lasso discard all variables with a higher \(\lambda\), exclusive lasso select one variable from each group. I add horizontal dotted lines for separating each group just for exposition purpose.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 | > df.comp.higher.lambda group beta Lasso Group Exclusive V1 1 0.150 0 0 0.0000000000 V2 1 -0.330 0 0 -0.0031930017 V3 1 0.250 0 0 0.0000000000 V4 1 -0.250 0 0 0.0000000000 V5 1 0.050 0 0 0.0000000000 ------------------------------------------- V6 2 0.000 0 0 0.0000000000 V7 2 0.000 0 0 0.0000000000 V8 2 0.000 0 0 0.0000000000 V9 2 0.500 0 0 0.0051059151 V10 2 0.200 0 0 0.0000000000 ------------------------------------------- V11 3 -0.250 0 0 -0.0026019669 V12 3 0.120 0 0 0.0000000000 V13 3 -0.125 0 0 0.0000000000 V14 3 0.000 0 0 0.0000000000 V15 3 0.000 0 0 0.0000000000 ------------------------------------------- V16 4 0.000 0 0 0.0000000000 V17 4 0.000 0 0 0.0000000000 V18 4 0.000 0 0 0.0006854416 V19 4 0.000 0 0 0.0000000000 V20 4 0.000 0 0 0.0000000000 |
This is interesting and may be useful when we want to select one security in each sector when forming a diversified asset portfolio with many investment sectors. Of course, a further analysis is necessary to select arbitrary predetermined number of securities from each sector.
Concluding Remarks
This post shows how to use group lasso and exclusive lasso using R code. In particular, I think that the exclusive lasso delivers some interesting result which will be investigated furthermore in following research such as sector-based asset allocation (sectoral diversification).
Reference
Yuan, M. and L. Lin (2006), Model Selection and Estimation in Regression with Grouped Variables, Journal of the Royal Statistical Society, Series B 68, pp. 49–67.
Zhou, Y., R. Jin, and S. Hoi (2010), Exclusive Lasso for Multi-task Feature Selection. In International Conference on Artificial Intelligence and Statistics, pp. 988-995.
Qiu, L., Y. Qu, C. Shang, L. Yang, F. Chao, and Q. Shen (2021), Exclusive Lasso-Based k-Nearest Neighbors Classification. Neural Computing and Applications, pp. 1-15. \(\blacksquare\)
Hello, thank you for your article!
ReplyDeleteMay I ask for my understanding, on which basis is the group index for Group LASSO selected?
Hi. group index is given using about three criteria.
Delete1) group such as sector or region of stocks (deterministic)
2) group by unsupervised learning or clustering (semi-endogenous)
3) group by one-hot encoding of categorical variables (technical)
I hope this helps your understanding. :-)
Thank you very much, that helped a lot already!
DeleteMany thanks for this insightful material!
ReplyDeleteI want to fit a LASSO Cox proportional hazards regression model for survival data. However, I'm having quite a hard time combining the failure (outcome) variable and the survival/follow-up time. I would really appreciate your help with that.
Tibshirani published kindly R package, which is your topic.
Deletehttps://cran.r-project.org/web/packages/glmnet/vignettes/Coxnet.pdf
Many thanks. I appreciate it.
Delete